Difference between revisions of "Examples of implementing a unique study ID"
(Created page with "==From INBC of University of Heidelberg== Claudia Pitzer, the core facility head, uses this explanation in a document handed out to the user: File naming: in order that each...") |
m |
||
| (14 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
==From INBC of University of Heidelberg== | ==From INBC of University of Heidelberg== | ||
| − | Claudia Pitzer, the core facility head, uses this explanation in a document handed out to the user: | + | [https://orcid.org/0000-0002-7663-1031 Claudia Pitzer], the core facility head of the [http://www.medizinische-fakultaet-hd.uni-heidelberg.de/Home.111344.0.html Interdisciplinary Neurobehavioral Core], uses this explanation in a document handed out to the user: |
| + | |||
File naming: in order that each data is unique and retrievable we suggest that you adhere to the following file naming convention: [date YYMMDD]-[the first letter of your first name together with your last name]-[free text]. If your surname is long you may use only the first 6 letters of it. | File naming: in order that each data is unique and retrievable we suggest that you adhere to the following file naming convention: [date YYMMDD]-[the first letter of your first name together with your last name]-[free text]. If your surname is long you may use only the first 6 letters of it. | ||
| − | + | ||
| − | + | ::: e.g. '''201108-cpitzer-catwalk with mice zQ98''' | |
| − | ==From Neuro-Behavioral-Analysis Unit of University of Lausanne== | + | |
| − | Leonardo Restivo, the core facility head, uses this nomenclature to store metadata at his core facility. | + | ==From Neuro-Behavioral-Analysis Unit, Department of Fundamental Neuroscience, University of Lausanne== |
| − | This system is only used to store metadata | + | [https://orcid.org/0000-0002-3709-5504 Leonardo Restivo], the core facility head of the [http://neurobau.com Neuro-BAU], uses this nomenclature to store metadata at his core facility.[[File:LRestivo_StudyID_210428.jpg|frame|]] |
| − | + | ||
| − | + | This system is only used to store metadata(project id, project info, experimental protocols) but it is not used for raw data. | |
| + | The experimental record ID is generated as follows: | ||
| + | |||
| + | Initials of the PI + Acronym of the department + Institution acronym + Sequential integer | ||
| + | |||
Example: | Example: | ||
| − | Leonardo Restivo, Department of Fundamental Neuroscience (swap letters in french acronym | + | Leonardo Restivo, Department of Fundamental Neuroscience (swap letters in french acronym), University of Lausanne, project one |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ::: e.g. '''LR_DNF_UNIL_1''' | ||
| + | This id is part of a simple two-table database (it has its unique own row_id see image): | ||
| + | The `code-name` field contains a friendly name (e.g. "gut microbiota and spatial orientation") for the project. This name is used to communicate with the lab working on the project. | ||
| + | Each experiment belonging to this project inherits the project id. Every experiment has its own unique id (which is a unique auto-incrementing integer) + friendly name (e.g. "water maze") | ||
| + | The `experiment` table has a field named `data`. This is a link to a text-markdown file that logs the experimental protocol and any change to it. | ||
| + | |||
==From Behavioral and Physiological Phenotyping Unit of Weizmann Institute of Science== | ==From Behavioral and Physiological Phenotyping Unit of Weizmann Institute of Science== | ||
| − | Michael Tsoory, the core facility head, describes the procudure for users as follows: | + | [https://orcid.org/0000-0003-0441-8757 Michael Tsoory], the core facility head at [http://www.weizmann.ac.il/vet/advanced-services/behavior-and-physiology/about Weizman Institute], describes the procudure for users as follows: |
| − | “The PI must communicate the IACUC Approval number to the CF and to the person(s) undertaking the Studies and Experiments. This number must be incorporated into the unique study-experiment ID by whomever assumes responsibility on maintaining the Experimental Records”. | + | ''“The PI must communicate the IACUC Approval number to the CF and to the person(s) undertaking the Studies and Experiments. This number must be incorporated into the unique study-experiment ID by whomever assumes responsibility on maintaining the Experimental Records”.'' |
| − | Here at the WIS, the IACUC# is the first and most important requirement; a PI CANNOT purchase lab animals without it (it is a “must fill in field” in the procurement system) NOR ask for a breeding procedure (similar “must fill in field in electronic form). | + | |
| − | Therefore a record keeping system may apply the below noted suggestion: | + | Here at the WIS, the IACUC# is the first and most important requirement; a PI CANNOT purchase lab animals without it (it is a “must fill in field” in the procurement system) NOR ask for a breeding procedure (similar “must fill in field in electronic form). |
| − | For example IACUC Approval 12340221-1 (IA) includes 3 studies (3 Study Protocols: SP1, SP2, SP3), one of which (SP2) is assessing the relevance of gene X to learning and memory. So SP2 includes assessing wt and KO mice in the 1) Y-maze; 2) NOR; 3) MWM; as well as some anxiety tests 4) Acoustic Startle Response; 5) Dark-Light Transfer test; and basic motor assessments 6) home-cage locomotion; 7) Open-Filed. All together 7 Experimental Procedures (EP). | + | |
| − | The Experimental Records will be filed in a FOLDER named: IA12340221-1 | + | Therefore a record keeping system may apply the below noted suggestion: |
| − | + | ||
| − | + | For example, '''IACUC Approval 12340221-1 (IA)'' includes 3 studies (3 Study Protocols: SP1, SP2, SP3), one of which ('''SP2''') is assessing the relevance of gene X to learning and memory. So SP2 includes assessing wt and KO mice in the 1) Y-maze; 2) NOR; 3) MWM; as well as some anxiety tests 4) Acoustic Startle Response; 5) Dark-Light Transfer test; and basic motor assessments 6) home-cage locomotion; 7) Open-Filed. All together 7 '''Experimental Procedures (EP)'''. | |
| − | + | ||
| + | The Experimental Records will be filed in a '''FOLDER named: IA12340221-1''' | ||
| + | * '''PLANS''', experimental designs, planed experimental procedures, and everything else that has to do with planning etc. (like calculations of Ns), will filed in folders named SP with subfolders for every experiment EP., | ||
| + | * All data generated in these studies will be filed a '''RAW DATA''' folder; a DATA folder will have subfolders of SP and subsequent EP. | ||
| + | * All analyses of data generated in these studies will be filed an '''ANALYSES''' folder | ||
(we may want to include additional folders for “conclusions”, “publications”) | (we may want to include additional folders for “conclusions”, “publications”) | ||
| − | IA12340221-1 | + | '''IA12340221-1''' |
| − | + | :'''PLANS''' | |
| − | + | ::SP1 | |
| − | + | ::SP2 | |
| − | + | :::EP1 | |
| − | + | :::EP2 | |
| − | + | :::… | |
| − | + | :::EP7 | |
| − | + | :'''RAW DATA''' | |
| − | + | ::SP1 | |
| − | + | ::SP2 | |
| − | + | :::EP1 | |
| − | + | :::EP2 | |
| − | + | :::… | |
| − | + | :::EP7 | |
| − | + | :'''ANALYSES | |
| − | + | ::SP1 | |
| − | + | ::SP2 | |
| − | + | :::EP1 | |
| − | + | :::EP2 | |
| − | + | :::… | |
| − | + | :::EP7 | |
The data generated in this MWM experiment will be | The data generated in this MWM experiment will be | ||
| − | e.g. IA12340221-1/DATA/SP2/EP3 | + | |
| + | e.g. '''IA12340221-1/DATA/SP2/EP3''' | ||
| + | |||
The files in that folder can take a name comprised of the DATE and initials of the person performing the test “John Doe”: JD_YYMMDD. | The files in that folder can take a name comprised of the DATE and initials of the person performing the test “John Doe”: JD_YYMMDD. | ||
| − | Moreover, in the data sheet I STRONGLY recommend that the IACUC#, SP# and EP# would appear in the columns next to the mouse#, DOB, SEX and Genotype columns (in the Noldus Ethovision these columns may be added already in the “trial list” stage and would be automated included in all output/export files): | + | Moreover, in the data sheet I STRONGLY recommend that the IACUC#, SP# and EP# would appear in the columns next to the mouse#, DOB, SEX and Genotype columns (in the Noldus Ethovision these columns may be added already in the “trial list” stage and would be automated included in all output/export files): |
| − | Identifiers | + | |
| − | Latency to reach platform | + | {|class="wikitable" |
| − | IACUC# | + | !colspan="7"|Identifiers |
| − | SP# | + | !colspan="4"|Latency to reach platform |
| − | EP# | + | |- |
| − | M# | + | |IACUC# |
| − | DOB | + | |SP# |
| − | SEX | + | |EP# |
| − | Genotype | + | |M# |
| − | Day1 Trail1 | + | |DOB |
| − | Day1 Trail2 | + | |SEX |
| − | ….. | + | |Genotype |
| − | Day5 Trail4 | + | |Day1 Trail1 |
| − | 12340221-1 | + | |Day1 Trail2 |
| − | 2 | + | |….. |
| − | 3 | + | |Day5 Trail4 |
| − | 666 | + | |- |
| − | YYMMDD | + | |12340221-1 |
| − | F | + | |2 |
| − | wt | + | |3 |
| − | 90 | + | |666 |
| − | 90 | + | |YYMMDD |
| − | + | |F | |
| − | 25 | + | |wt |
| − | 12340221-1 | + | |90 |
| − | 2 | + | |90 |
| − | 3 | + | | |
| − | 667 | + | |25 |
| − | YYMMDD | + | |- |
| − | m | + | |12340221-1 |
| − | + | |2 | |
| − | 90 | + | |3 |
| − | + | |667 | |
| − | + | |YYMMDD | |
| − | 34 | + | |m |
| − | + | |vKO | |
| − | 2 | + | |90 |
| − | 3 | + | |v87 |
| − | 668 | + | | |
| − | YYMMDD | + | |34 |
| − | F | + | |- |
| − | KO | + | |v12340221-1 |
| − | 90 | + | |2 |
| − | 90 | + | |3 |
| − | + | |668 | |
| − | 18 | + | |YYMMDD |
| − | 12340221-1 | + | |F |
| − | 2 | + | |KO |
| − | 3 | + | |90 |
| − | 669 | + | |90 |
| − | YYMMDD | + | | |
| − | m | + | |18 |
| − | wt | + | |- |
| − | 90 | + | |12340221-1 |
| − | 78 | + | |2 |
| − | + | |3 | |
| − | 27 | + | |669 |
| − | + | |YYMMDD | |
| + | |m | ||
| + | |wt | ||
| + | |90 | ||
| + | |78 | ||
| + | | | ||
| + | |27 | ||
| + | |} | ||
Latest revision as of 07:51, 29 April 2021
From INBC of University of Heidelberg
Claudia Pitzer, the core facility head of the Interdisciplinary Neurobehavioral Core, uses this explanation in a document handed out to the user:
File naming: in order that each data is unique and retrievable we suggest that you adhere to the following file naming convention: [date YYMMDD]-[the first letter of your first name together with your last name]-[free text]. If your surname is long you may use only the first 6 letters of it.
- e.g. 201108-cpitzer-catwalk with mice zQ98
From Neuro-Behavioral-Analysis Unit, Department of Fundamental Neuroscience, University of Lausanne
Leonardo Restivo, the core facility head of the Neuro-BAU, uses this nomenclature to store metadata at his core facility.
This system is only used to store metadata(project id, project info, experimental protocols) but it is not used for raw data. The experimental record ID is generated as follows:
Initials of the PI + Acronym of the department + Institution acronym + Sequential integer
Example: Leonardo Restivo, Department of Fundamental Neuroscience (swap letters in french acronym), University of Lausanne, project one
- e.g. LR_DNF_UNIL_1
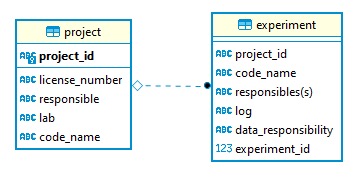
This id is part of a simple two-table database (it has its unique own row_id see image): The `code-name` field contains a friendly name (e.g. "gut microbiota and spatial orientation") for the project. This name is used to communicate with the lab working on the project. Each experiment belonging to this project inherits the project id. Every experiment has its own unique id (which is a unique auto-incrementing integer) + friendly name (e.g. "water maze") The `experiment` table has a field named `data`. This is a link to a text-markdown file that logs the experimental protocol and any change to it.
From Behavioral and Physiological Phenotyping Unit of Weizmann Institute of Science
Michael Tsoory, the core facility head at Weizman Institute, describes the procudure for users as follows:
“The PI must communicate the IACUC Approval number to the CF and to the person(s) undertaking the Studies and Experiments. This number must be incorporated into the unique study-experiment ID by whomever assumes responsibility on maintaining the Experimental Records”.
Here at the WIS, the IACUC# is the first and most important requirement; a PI CANNOT purchase lab animals without it (it is a “must fill in field” in the procurement system) NOR ask for a breeding procedure (similar “must fill in field in electronic form).
Therefore a record keeping system may apply the below noted suggestion:
For example, IACUC Approval 12340221-1 (IA) includes 3 studies (3 Study Protocols: SP1, SP2, SP3), one of which ('SP2) is assessing the relevance of gene X to learning and memory. So SP2 includes assessing wt and KO mice in the 1) Y-maze; 2) NOR; 3) MWM; as well as some anxiety tests 4) Acoustic Startle Response; 5) Dark-Light Transfer test; and basic motor assessments 6) home-cage locomotion; 7) Open-Filed. All together 7 Experimental Procedures (EP).
The Experimental Records will be filed in a FOLDER named: IA12340221-1
- PLANS, experimental designs, planed experimental procedures, and everything else that has to do with planning etc. (like calculations of Ns), will filed in folders named SP with subfolders for every experiment EP.,
- All data generated in these studies will be filed a RAW DATA folder; a DATA folder will have subfolders of SP and subsequent EP.
- All analyses of data generated in these studies will be filed an ANALYSES folder
(we may want to include additional folders for “conclusions”, “publications”)
IA12340221-1
- PLANS
- SP1
- SP2
- EP1
- EP2
- …
- EP7
- RAW DATA
- SP1
- SP2
- EP1
- EP2
- …
- EP7
- ANALYSES
- SP1
- SP2
- EP1
- EP2
- …
- EP7
The data generated in this MWM experiment will be
e.g. IA12340221-1/DATA/SP2/EP3
The files in that folder can take a name comprised of the DATE and initials of the person performing the test “John Doe”: JD_YYMMDD. Moreover, in the data sheet I STRONGLY recommend that the IACUC#, SP# and EP# would appear in the columns next to the mouse#, DOB, SEX and Genotype columns (in the Noldus Ethovision these columns may be added already in the “trial list” stage and would be automated included in all output/export files):
| Identifiers | Latency to reach platform | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IACUC# | SP# | EP# | M# | DOB | SEX | Genotype | Day1 Trail1 | Day1 Trail2 | ….. | Day5 Trail4 |
| 12340221-1 | 2 | 3 | 666 | YYMMDD | F | wt | 90 | 90 | 25 | |
| 12340221-1 | 2 | 3 | 667 | YYMMDD | m | vKO | 90 | v87 | 34 | |
| v12340221-1 | 2 | 3 | 668 | YYMMDD | F | KO | 90 | 90 | 18 | |
| 12340221-1 | 2 | 3 | 669 | YYMMDD | m | wt | 90 | 78 | 27 | |